Molecular Ligand Target Research Team
- Main Research Fields :
- Biology
- Related Research Fields :
- Medicine, Dentistry & Pharmacy
- Keywords :
- Chemical Genomics / Target Identification / Yeast
- Project :
- Advanced Research and Technology Platforms
TP
Exploring target molecules and mode-of-action of bioactive compounds through global analysis of chemical genetic interactions
Team Leader
Charles M. Boone Ph.D.

- 1989
- Ph.D., McGill University, Canada
- 1993
- Assistant Professor, Simon Fraser University, Canada
- 1997
- Assistant Professor, Queen's University, Canada
- 2000
- Associate Professor, University of Toronto, Canada
- 2003
- Professor, University of Toronto, Canada
- 2008
- Senior Visiting Scientist, RIKEN
- 2009
- Team Leader, Molecular Ligand Target Research Team, RIKEN
- 2013
- Team Leader, Molecular Ligand Target Research Team, RIKEN Center for Sustainable Resource Science (-current)
CONTACT
RIKEN Center for Sustainable Resource Science
Chemical Biology Research Group,
Molecular Ligand Target Research Team
charlieboone
Related Links
Laboratory Website
Molecular Ligand Target Research Team
Laboratory on RIKEN Website
Molecular Ligand Target Research Team | RIKEN
Outline

Subjects
- Global analysis of chemical genetic interactions between molecular ligands and their target molecules
- Validating the mode of action of bioactive compounds
- Identifying bioactive chemical tools and therapeutic leads that target essential gene pathways
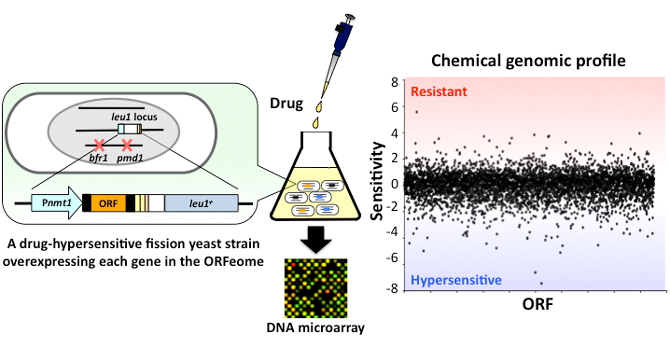
- A microarray-based method for identification of drug targets using the fission yeast ORFeome
- We created a novel strain collection in a drug-sensitive genetic background that expresses the entire fission yeast ORFeome. We can now pool this library and grow the yeast in the presence of a drug. The abundance of each strain is assessed using DNA microarray to generate a genome-wide profile of drug-gene interactions. This chemical genomics approach can rapidly determine genes involved in pathways targeted by small molecules.





