Metabolic Systems Research Team
- Main Research Fields :
- Biological Sciences
- Related Research Fields :
- Biology / Agricultural Sciences
- Keywords :
- Metabolism / Metabolomics / Mathematical modelling / Plant / Secondary metabolism
- Project :
- Metabolic Genome Engineering / Advanced Research and Technology Platforms
M
TP
Understanding the mechanisms and physiology of plant metabolism and improving production of useful materials
Team Leader
Masami Yokota Hirai Ph.D.

- 1994
- Ph.D., Agriculture, University of Tokyo
- 1994
- JSPS Postdoctoral Fellow, University of Tokyo
- 1997
- Research Associate, Chiba University
- 2001
- CREST Postdoctoral Fellow, Japan Science and Technology Agency
- 2005
- Unit Leader, Metabolic Systems Research Unit, RIKEN Plant Science Center
- 2005
- Adjunct Associate Professor, School of Agricultural Sciences and Graduate School of Bioagricultural Sciences, Nagoya University
- 2006
- Visiting Associate Professor, University of Tokyo
- 2007
- Adjunct Professor, Nagoya University (-current)
- 2008
- Team Leader, Metabolic Systems Research Team, RIKEN Plant Science Center
- 2010
- Visiting professor, Alkali Soil Natural Environmental Science Center, Northeast Forestry University, China
- 2013
- Team Leader, Metabolic Systems Research Team, RIKEN Center for Sustainable Resource Science (-current)
- 2020
- Unit Leader, Mass Spectrometry and Microscopy Unit, RIKEN Center for Sustainable Resource Science (-current)
CONTACT
RIKEN Center for Sustainable Resource Science
Metabolic Systems Research Team
masami.hirai
YokohamaAccess
1-7-22 Suehiro, Tsurumi, Yokohama, Kanagawa 230-0045 Japan
#C619 6F Central Research Building
Related Links
Laboratory Website
Metabolic Systems Research Team
Laboratory on RIKEN Website
Metabolic Systems Research Team | RIKEN
Outline

Subjects
- Elucidation of the regulatory mechanism of amino acid biosynthesis
- Identification of genes involved in biosynthesis/degradation of plant specialized metabolites
- Identification of metabolic pathways regulating plant development
- Data mining from metabolome data through machine learning and mathematical modeling
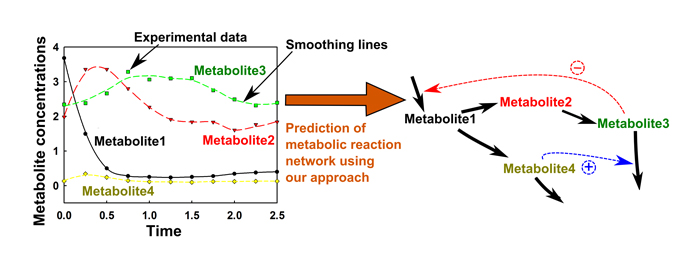
- Image of prediction of metabolic reaction network based on time-series metabolite data
- We established a novel algorithm for prediction of metabolic reaction network. The algorithm is comprised of 4 steps: smoothing of time-series metabolite data, determination of causal relationship by Granger causality test, modeling of metabolic reaction network by biochemical system theory, and estimation of parameters by nonlinear least-square method. (Sriyudthsak et al. PLOS ONE 2013)
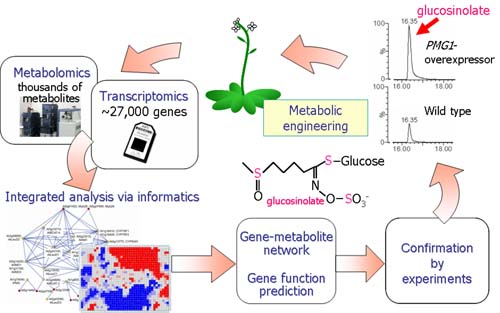
- Identification of a key transcription factor regulating glucosinolate biosynthesis
- Based on coexpression analysis using the transcriptome data, we identified candidate genes involved in glucosinolate biosynthesis, and confirmed the predicted function of the candidates by widely targeted metabolomics. (Hirai et al. PNAS 2007)





